John Walk
Empath Engine (Insight Data Science project)
John Walk
About Insight Data Science
Insight Data Science is an intensive postdoctoral fellowship geared towards STEM PhDs making the transition to data science in industry. The fellowship is centered around an individual demo product – each fellow spends 3-4 weeks building a tech demonstration solving a real-world data problem from conceptualization all the way through to a self-contained presentation (typically in the form of a webapp for the product). This demo is then presented to the sponsor companies over the following three weeks.
Rather than focusing on the technical aspects of the product (since, by and large, fellows enter the program already familiar with technical DS tools), Insight primarily focuses on the product side – project conceptualization and effective presentation to broad (frequently nontechnical) audiences. Bluntly, a project with the most cutting-edge tech out there doesn’t mean much if it’s presenting a solution to a problem no one cares about in a way that no one can follow.
The Empath Engine
For my Insight project, I built the empath engine – a socially-driven recommender and sentiment reporter for antidepressant medications.
The core problem is simple: while antidepressant prescription is extremely common (with some one in eight Americans currently taking antidepressants), the combination of social stigma and depressive symptoms themselves can make open discussion of symptoms, treatments, and medications difficult. Moreover, there is significant trial and error involved in antidepressant prescription, with numerous patients cycling through several different medications before settling on one. In short, the tendency towards isolation imposed by the disease interferes exactly where open discussion of others’ experiences would be helpful.
However, it seems that if you just add some anonymity via typical social media channels into the mix, people will much more freely discuss their experiences. I used data from reddit for this project, as it is suitably high traffic, anonymized behind persistent usernames, and supplies rich text data (comments capped at 10,000 characters, providing much more thorough information than e.g. Twitter) along with extensive metadata (timing, connections to other comments/posts) via an open API.
The Data
Reddit provides access to all public comment and post data via an open API. However, this is geared more towards live tool development (e.g., comment bots or feed streams) rather than after-the-fact data mining – in particular, the API is limited to two calls per second, with each call returning a maximum of 1000 query results due to upstream data structure limitations. Fortunately, a reddit user (conveniently found through /r/datasets) had already assembled these into a dataset by sequentially running API calls over the course of around 10 months(!) daisy-chaining the API results together via the sequential IDs associated with each post, hosting the result at pushshift.io.
Given this source, it is a fairly straightforward problem to find comments mentioning antidepressants. However, Reddit currently receives up to two million comments per day, with each monthly data dump reaching into the tens of gigabytes… of which less than a hundredth of a percent were relevant for this project. This necessitated streaming & out-of-core computing, which at the time I had little experience with – speed of data acquisition was my first major hurdle. Even so, in my limited time frame I assembled a set of some 30,000 relevant comments, indexed by the mentioned medication(s).
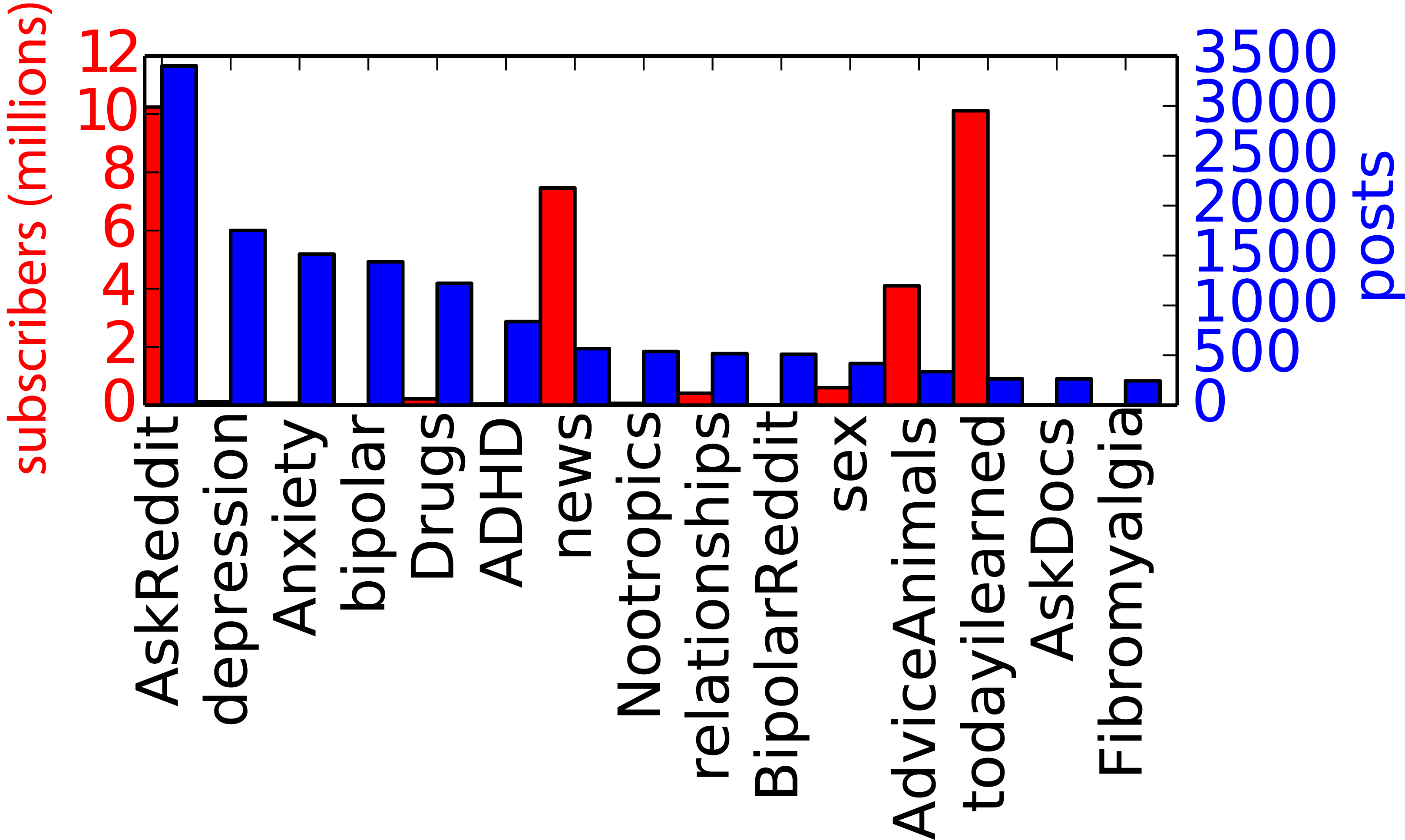
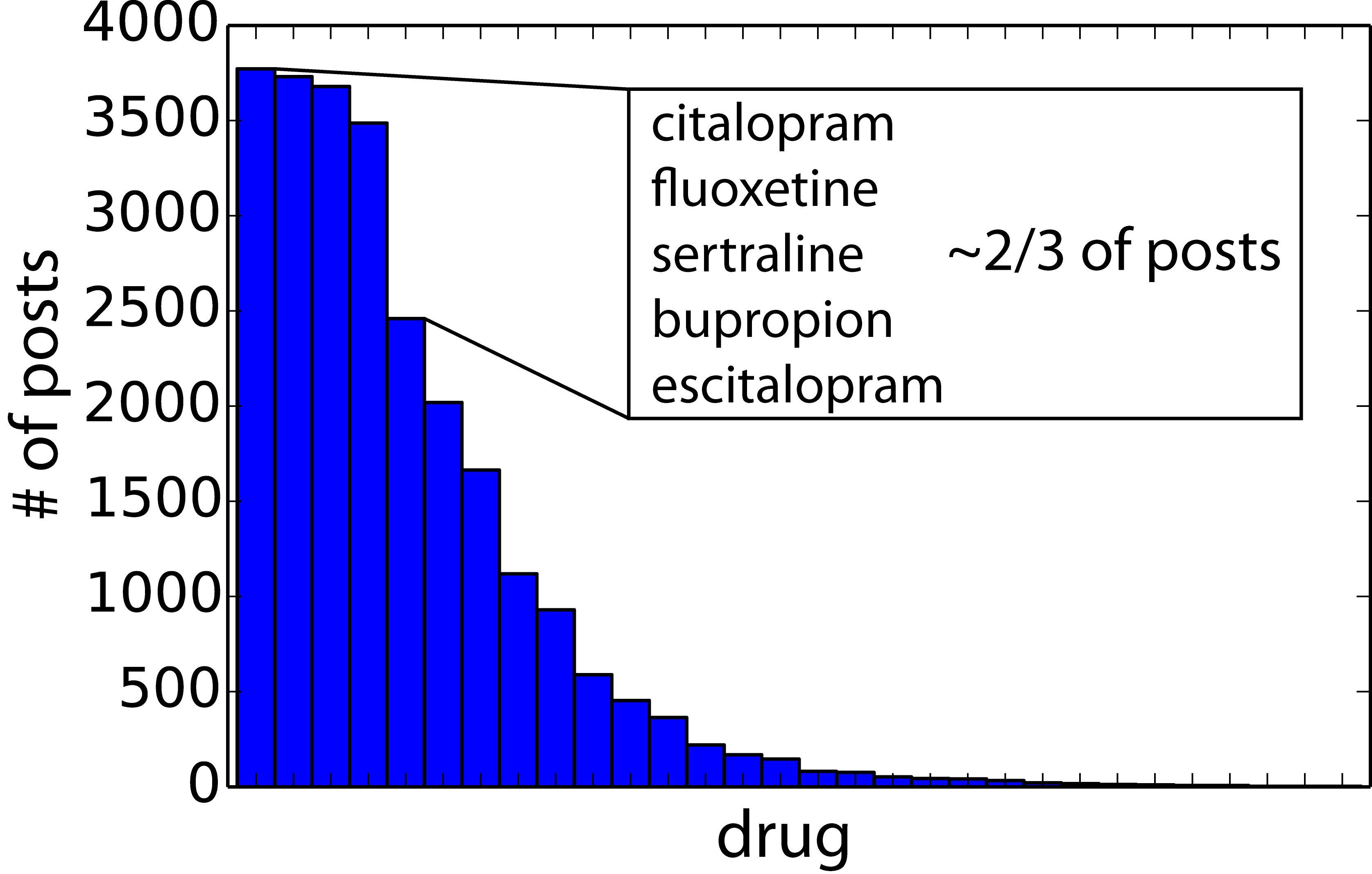
These came from a variety of source “subreddits” (user-driven topical sub-forums on reddit) – both from (a) a low “signal to noise” percentage of relevant posts coming from overall high-traffic subs (like /r/AskReddit) and (b) a high percentage of relevant traffic from smaller, topically-specific subs (e.g. /r/depression). While every medication recognized by the FDA for antidepressant usage was contained in the dataset, the bulk of the dataset was comprised of the most commonly prescribed drugs:
- citalopram (celexa)
- fluoxetine (prozac)
- sertraline (zoloft)
- bupropion (wellbutrin)
- escitalopram (lexapro)
Topic Modeling
For the first pass at analysis, I built a medication-specific topic model from the comments using term frequency-inverse document frequency (TF-IDF) scoring. First, each comment text (“document” in typical natural-language processing parlance) was preprocessed:
- obvious low-quality comments removed (deleted comments, reported spam, etc.)
- trade names for drugs remapped to consistent generics
- comments tokenized into discrete word/phrase entities
- “stop words” removed: conjunctions, articles, helper verbs, plus a hand-tuned list of other low-information tokens
- lemmatization, removing grammatical variation (e.g., “drug” and “drugs” map to the same token)
For a single document, it is trivial to reflect term importance by token frequency – that is, we use the assumption that the more a given term is used, the more central to the document topic it is. However, this will capture words that are common in similar documents, as well as words that are simply semantically common (as stop-word filtering cannot catch all of these). Instead, it is typical to weight the term frequency by inverse document frequency, which measures how often the term occurs in other documents: formally, for term \(t\) and document \(d\) from set \(D\),
\[idf(t,D) = \log \frac{N}{|{d \in D; t \in d}|}\]Describing the ratio of the number of documents containing the term \(|{d \in D; t \in d}|\) to the total size of the document corpus \(N = |D|\), inverted and log-scaled. This has the effect of downweighting terms that are common in general – essentially, a word common across most documents will have its naturally high term frequency offset by a low (approaching \(\log(1) = 0\) for truly universal terms) IDF.
In standard practice, this is done on a per-document basis out of a consistent corpus – for example, the TF of a term for a given news article is weighted by the IDF of that term from other news articles in the same source, allowing for article-specific keyword & topic tagging. However, given my corpus of drug-related comments, this approach fell short: I wanted to find terms that both distinguished the medication comments from general conversation (isolating medically-relevant terms) and distinguished medications from each other as much as possible. Rather, I treated each drug’s subset of the corpus as a single aggregate document (since individual comments were too short to provide meaningful statistics due to data sparsity) for calculating term frequency, and compared this to an IDF generated from the included web text corpus collated in the NLTK Python package. In effect, this targeted terms that were frequent in aggregate over comments for each drug, but not common in webforum traffic in general. Although it did require some hand-tuning for additional stopwords, this approach found fairly fine-grained results – for example, the use of Wellbutrin (Bupropion) off-label as a smoking cessation aid was captured from the comments.
Semantic Parsing & Sentiment Analysis
Next, I approached sentiment analysis (gauging positive or negative sentiment from post texts) for each drug. Typically this is a supervised task, requiring training texts pre-labeled with positive or negative sentiment – fortunately, I was able to supplement the training with a labeled comment dataset from AskaPatient scraped by a previous Insight fellow to build this labeler.
For a first pass, I built a Naive Bayes classifier – simply put, the classifier defines a probability of a class label \(C\) based on a vector \(\vec{x}\) of inputs (in this case, the tokens of a preprocessed text) via
\[p(C|\vec{x}) = \frac{p(\vec{x}|C)p(C)}{p(\vec{x})} \propto p(\vec{x}|C)p(C)\]Via Baye’s Theorem. The simplifying “naive” assumption comes by taking each token \(x_i \in \vec{x}\) as independent of all the others, allowing us to ignore relationships between them:
\[p(C|\vec{x}) \propto p(C)p(x_1|C)p(x_2|C)...p(x_n|C)\]where calculating each \(p(x_i|C)\) simply requires tallying counts from the training data. This allows extremely efficient computation even on very high-dimensional data like tokenized text. However, the assumption of independence between individual words in natural language is well… rather poor. For one, it cannot capture even very simple semantic information outside of individual words – without additional heuristics, a Naive Bayes classifier won’t necessarily even be able to distinguish “excellent” and “not excellent”! While this classifier was useful for assigning scores to individual terms from the TF-IDF-driven keyword set, a more fine-grained approach was needed for the reddit comments themselves.
To that end, I used the sentiment tool built in Stanford’s CoreNLP package, which operates by (a) building a parse tree mapping the semantic structure of the sentence and (b) using a recursive neural network to assign a sentiment label at each node of the tree.
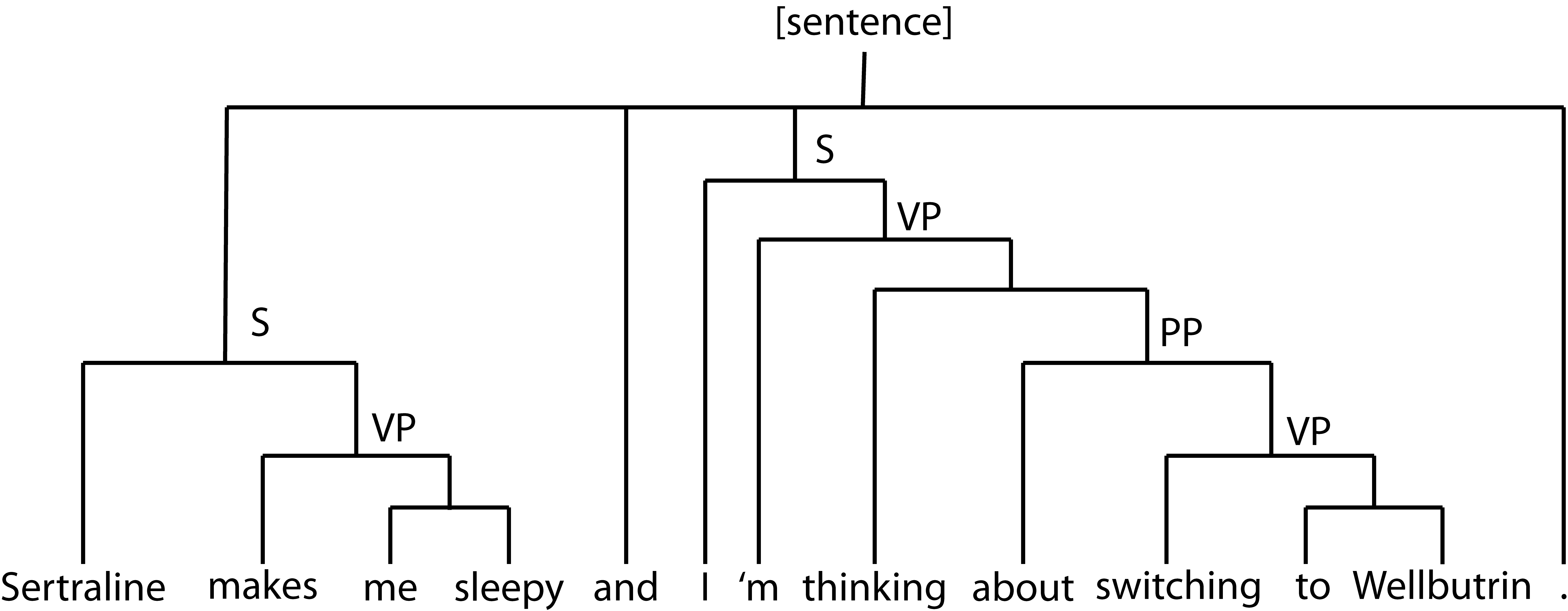
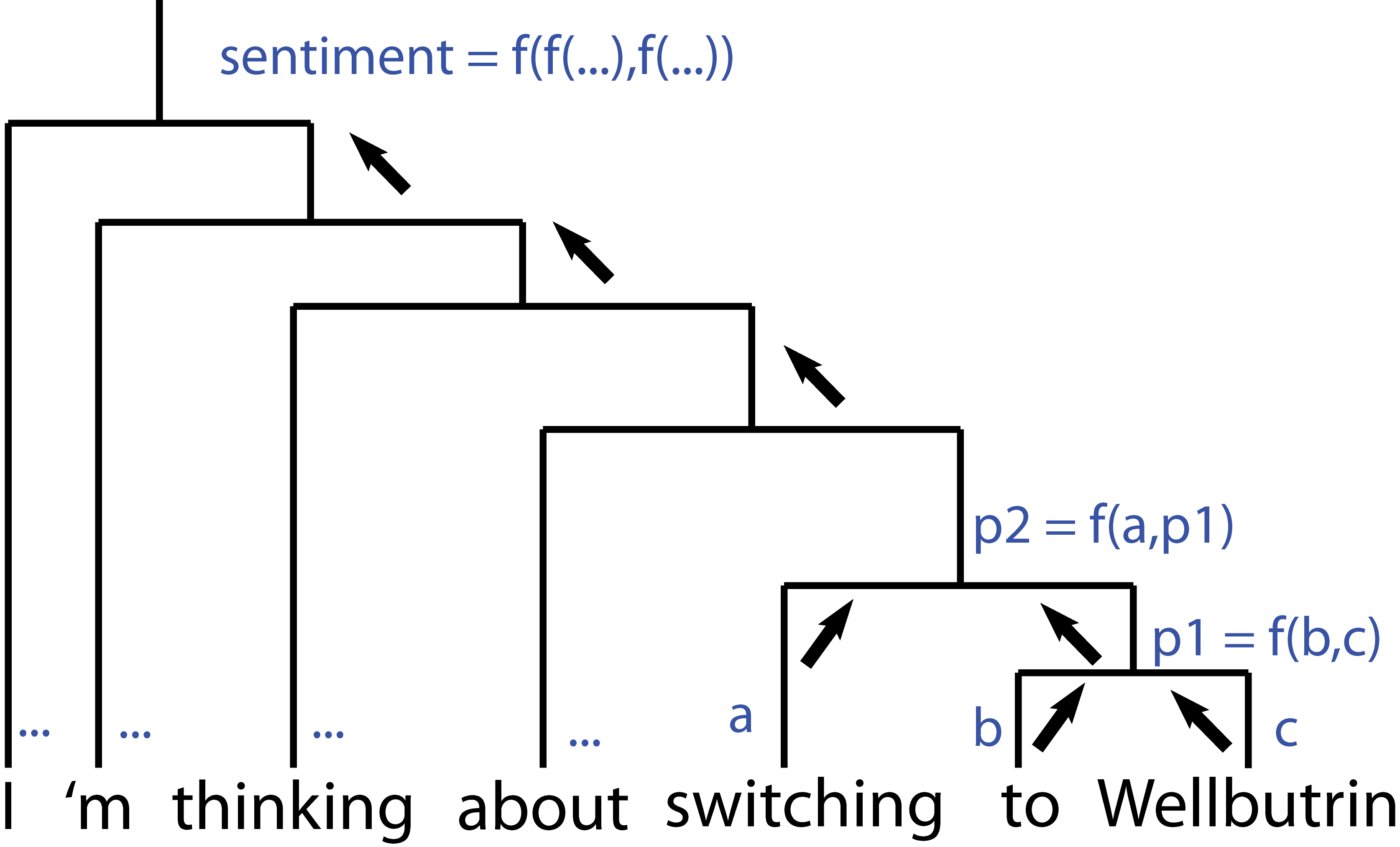
In addition to generating sentiment scores (via a pre-trained sentiment analyzer included in the tool, trained on Stanford’s combined dataset of parsed and labeled general text) accounting for semantic information, this let me intelligently separate components of comments talking about different medications – the sentence “chunks” were simply the maximum-height subtrees containing exactly one drug mention.
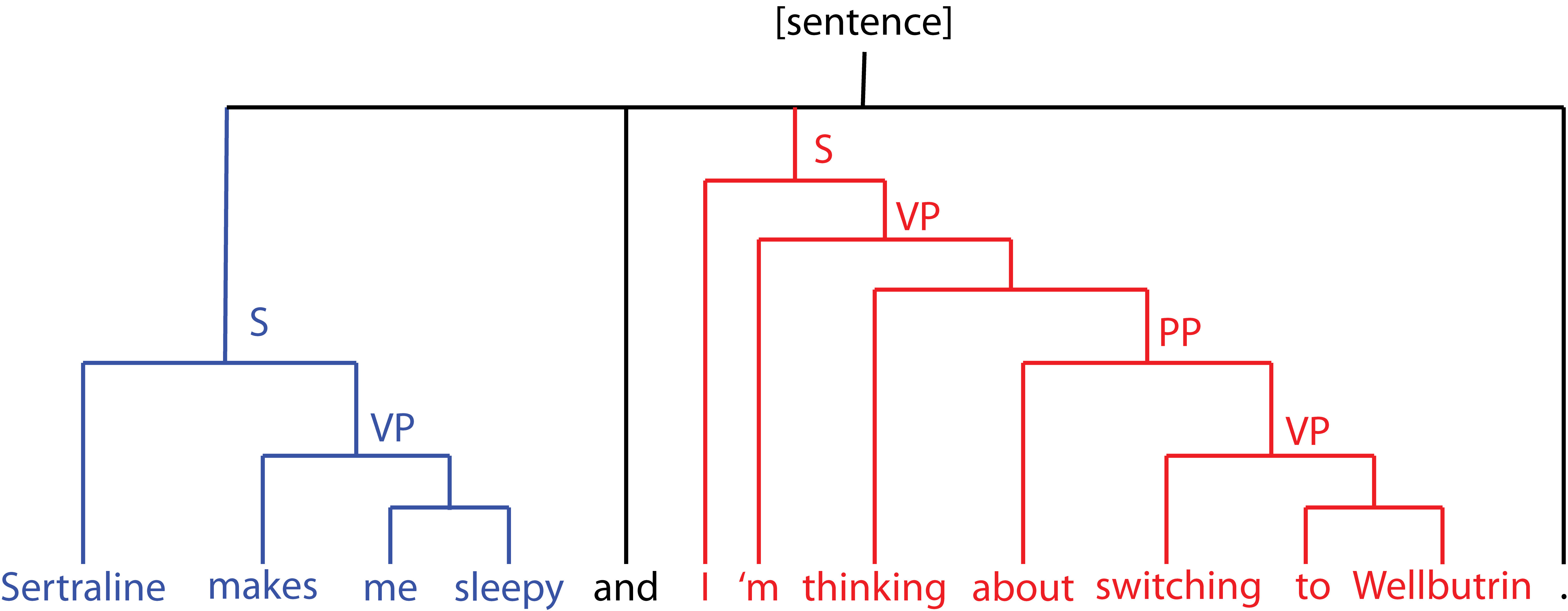
Since the neural network assigns sentiment labels at each node (recursing these up the tree to generate the overall sentiment) I also had a sentiment score natively assigned to these same subtrees.
The Recommender
Given drug-specific sentiment labeling, I built a simple reporter and recommender app. For a given drug, the app reports overall sentiment scores, as well as topic-specific keywords annotated with Naive Bayes-generated sentiment labels. Naturally, sentiment talking about experiences with antidepressants tends to skew negative – thus the drug sentiment is measured compared to a baseline across the drug-related corpus.
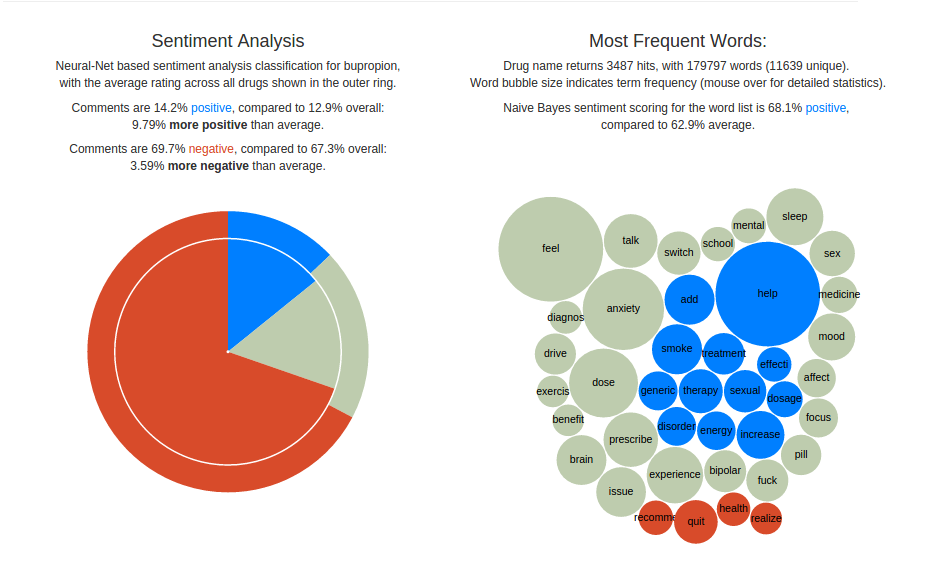
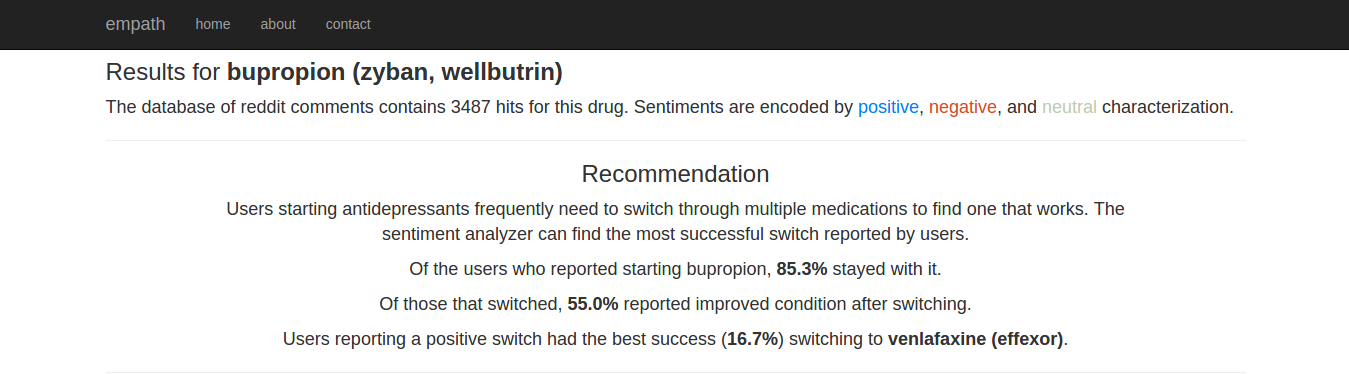
Based on any comments mentioning multiple drugs, the app would also make recommendations using the differential in sentiment score. That is, based on the assumption that descriptions were largely chronological (“drug A, then drug B”), for each drug A the corresponding B with the greatest positive differential in sentiment labeling was recommended, along with descriptive statistics.
Lessons Learned
This project was my first foray into natural language processing, and it was certainly a fascinating rabbit hole – it seems that NLP can be basically unbounded in the level of complexity one can apply to a task. I found it beneficial to strictly bound the complexity of solutions I would apply for the MVP of the recommender, either by using prepackaged tools (like the pre-trained CoreNLP sentiment analysis) or by making simplifying assumptions to the structure of the data to allow for more regular processing rather than getting lost in the edge cases (which are especially numerous for NLP).
Insight projects are frequently an exercise in how quickly one can create a minimum viable product – indeed, at numerous points during the process I would think of improvements to the previous week’s work and have to resist the temptation to go back to make fixes on (sufficiently working) components. A few improvements that could be made:
-
fast processing of the raw data – parsing the raw reddit comments was quite time consuming due to the small number of relevant comments compared to total traffic. However, as the comments were distributed as monthly dumps, the filtering task was embarassingly parallelizable, and presented numerous opportunities for speed-up. The final size of my corpus was limited by this speed, as I was working under time constraints, so more data could have been acquired – but I was limited by my lack of experience (at the time) with parallel/distributed computing tools.
-
better integration with the CoreNLP tools – while the data preparation and preprocessing was done entirely in Python (largely using the standard NLTK package), CoreNLP is natively distributed in Java. The corresponding Python API lagged behind the development of the core tools and presented several compatibility issues, as well as a time-consuming learning curve. Instead, I simply ran the CoreNLP tools as compiled binary calls triggered from the Python scripts, with the results written to XML files that were then re-read into Python (itself requiring some parser hacks). This admittedly was a “Frankencode” solution that was inefficient both in terms of complexity and computational speed. Better integration of the parser tooling into the rest of the pipeline would likely be the single greatest improvement to the data product possible.