John Walk
Scalable Data Processing Pipelines with Open-Source Tools
John Walk
this presentation was developed while at Cinch Financial.
In my experience at Cinch, building a complete picture of users’ financial lives required integrating user-supplied answers, credit data & linked accounts, 3rd-party vendor data, and open-access datasets into a single unified picture. This then needs to be made available for predictive modeling and analytics in a scalable, reproducible way, independent of live production data – both to divorce R&D development from the rest of the software cycle, and to ensure efficient access and security.
understanding the problem
The beginning and end of the data process (for any data-driven organization, really) are straightforward. On the one side, we have our sources: user-supplied answers, web/behavioral data, 3rd-party vendor data, or government/open-source data. Likewise, for the goals, we want user analytics, validation at scale of the advice we’re offering, and data-driven intelligence and ML models.
In between these, though…

here there be dragons.
When I joined Cinch, analytics and intelligence development followed a rather old-school paradigm:
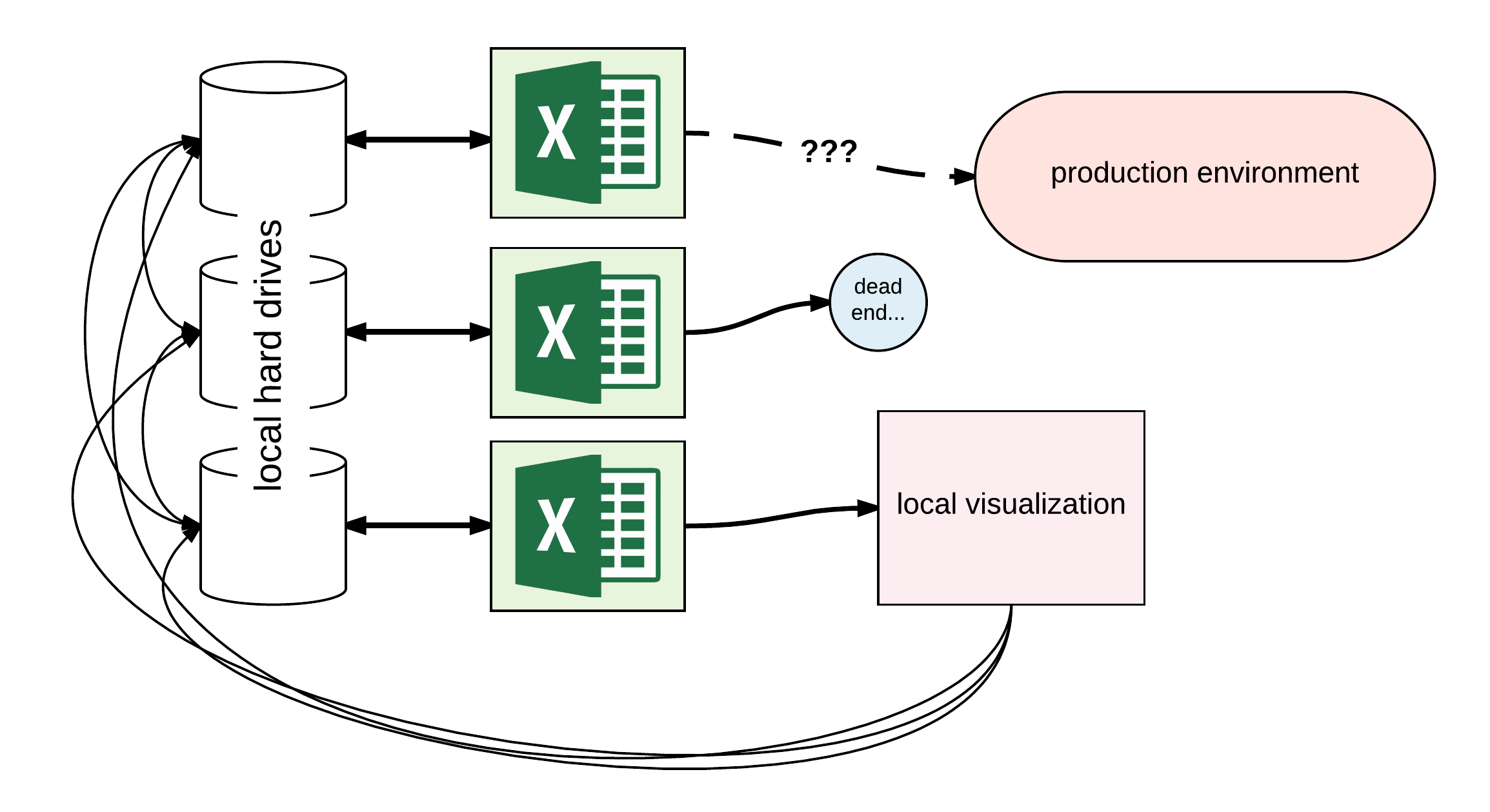
Data was local on analysts’ laptops or shared via web drop points (e.g. Google Drive), analysis and visualization was done locally in Excel, and the pathway for integration with the production environment required re-implementing business logic into a machine-readable form. Sharing analysis went through a tangled web of emails and web drop-points, often ending up in dead ends. Even preprocessed data had versioning issues since local analyses could get out of sync with each other relative to the central store (since it wasn’t really central data access at all, but rather a static box). Despite being a pre-launch company without large volumes of user data, the BI team when I joined was already out of the scale that these tools could handle.
Clearly, a new approach was needed – as part of revamping the intelligence products team at Cinch, I wanted to build out a scalable, flexible data analysis pipeline end-to-end from raw data to production-ready insight and modeling.
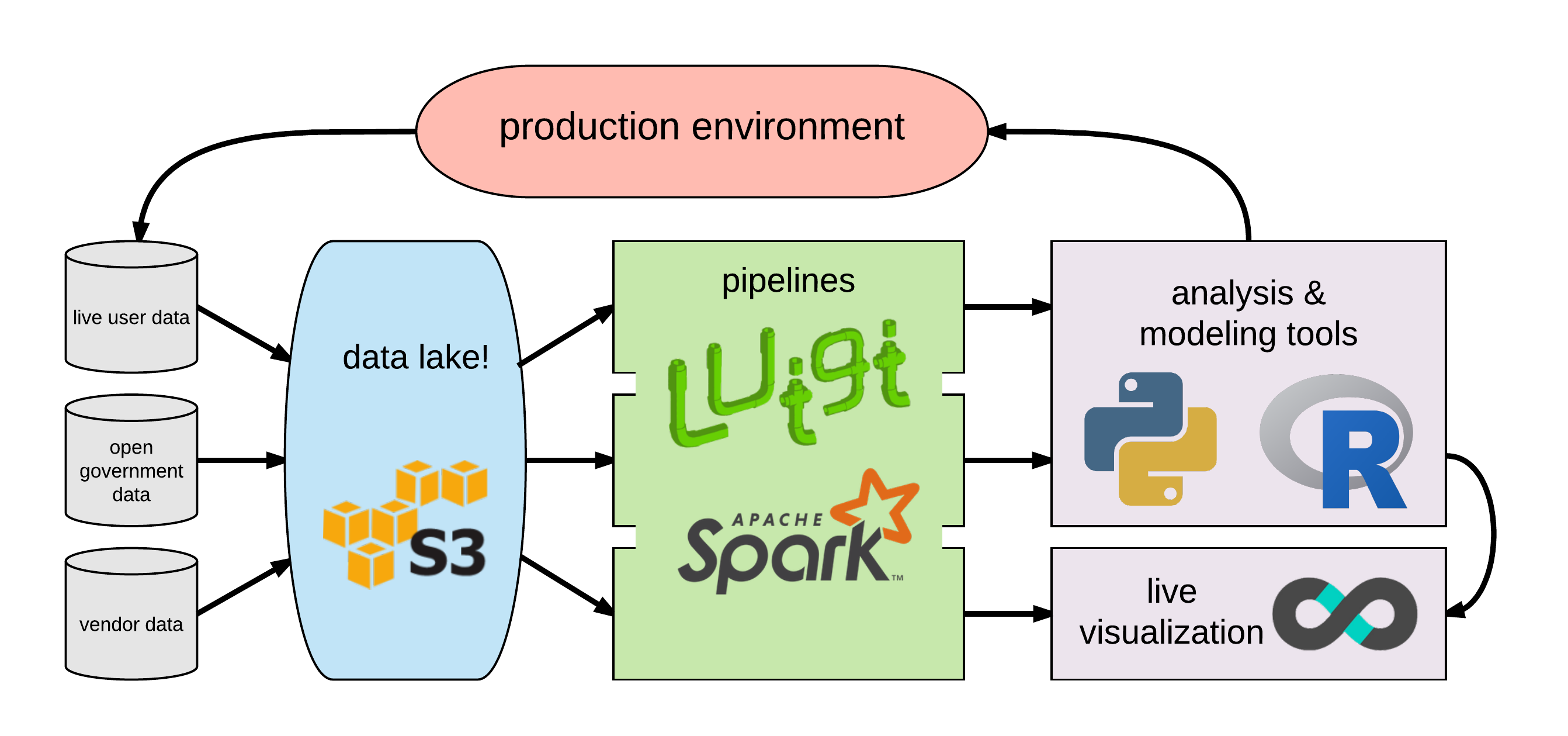
| “data lake” storage | unified access for DS, analysts |
| data pipelines | reproducible, fault-tolerant, modular ETL processing |
| scalable analysis tools | shared resources & tools for analysis/modeling on large data volume |
| visualization tools | accessible leverage on scaled data |
This meant a ground-up redesign of how we handled data storage, ETL processing, tooling for analysis & modeling, and shareable visualization tools/dashboards. Each of these exist as commercial products: for example, Databricks for storage/ETL or Tableau for visualization. However, with a little cleverness it’s possible to build this pipeline entirely with free and open-source tools – since my grad work had made extensive use of (and some contributions to) open-source tools, this definitely piqued my interest.
the data lake
ok, so what is it?
Simply put, a data lake is an unstructured storage system of data held in its original format – CSVs, JSONs, even image files. This should satisfy the same needs as an OLAP database (frequently called a “data warehouse”) – however, rather than trying to design a unified schema to fit all your data into, you let the just-in-time processing by the pipeline take your data from its raw form to the shape needed for analytics use. The goal is to avoid biasing your data – by leaving it in its original state, you retain maximum information from its source. Bluntly, when you try to optimize your data structure to answer certain questions, those tend to become the only questions you ask it.
In many applications, fitting data into a rigid schema isn’t really an problem: sales records for an online retailer will generally always look the same structurally, for example. However, when dealing with an application that may require integrating new data sources and applications (e.g., opening a new financial vertical at Cinch), then having a static data schema can force either (a) rebuilding substantial portions of the data system or (b) bizarre monkey-patches to force the new source to fit the existing schema.
Of course, this approach is not without its pitfalls – a data lake can easily become a “data swamp” with unstructured data lacking any coherence, making it near-impossible to extract useful insight from it. The key to avoiding this is ownership, documentation, knowledge transfer: the more analysts can have complete mastery over a component of the lake (and document and pass on their expertise), the easier it is to make sure your data lake remains useful.
Fortunately, implementing a data lake is quite simple: you just need simple, scalable file storage. We used Amazon S3 as a simple bootstrap setup, since it is
- trivial to set up
- relatively cheap (per unit storage)
- has interfaces to other major tools
- is infinitely scalable (just pay for more space)
However, S3 is emphatically not a filesystem, nor is it optimized for live data access. This can be mitigated with a good download pipe (or if your other data tools are also hosted on AWS resources). If more scalable resources are needed, AWS also offers Elastic Block Storage (EBS) or Elastic Filesystem (EFS) options, or one can host a Hadoop Distributed Filesystem (HDFS) instance on one’s own resources.
the data pipeline
wait, isn’t this just a script?
For simple jobs, sure – all you need is a runnable script to process data from its raw form in the lake to whatever shape is needed for analytics or modeling. If you’re clever, you can even package this into a CLI tool or executable for non-coder teammates.
but what about complex jobs?
As the complexity of your data process grows, simple scripts or CRON jobs start to fall short in three ways:
-
fault tolerance: if anything fails, the whole script breaks – and this compounds as you integrate more diverse tools and data formats
-
reproducibility: once you have a recipe for a particular task, why not reuse it (ideally without copy-pasting code?
-
scalability: you can learn to write good parallel code… but if you’re just doing a lot of largely-independent tasks then that’s overkill
enter the task graph
Instead of a monolithic script, it’s helpful to envision the data pipeline as a graph, with each node in the graph corresponding to an ETL task, and each edge encoding a dependency. From a a computational standpoint, we specifically want a directed acyclic graph (DAG) – this just means that the connections are directional (so A-B is not the same as B-A between nodes) and that the graph has no loops, so will always get to an end point. In terms of our failure modes above, this is:
-
fault-tolerant, since failed tasks just pause that path on the graph – the graph continues to execute other paths as long as dependencies are met. The graph also remembers its state, so it can be resumed without repeating work.
-
reproducible, in that each node encodes a reusable recipe for an atomic task
-
scalable, since independent tasks can be executed by multiple workers in parallel
Luigi – the world’s second-most famous plumber
For this task, I like Luigi, a Python package for ETL tasks open-sourced by the DS team at Spotify.
Luigi is designed to need minimal boilerplate to specify the task graph. Each
task node type is laid out as a subclass of luigi.Task, and only requires
three basic methods: requires to define the upstream dependencies, output
to specify the output location (a local or remote filesystem, or a DB
connection), and run to execute arbitrary Python code for the task itself.
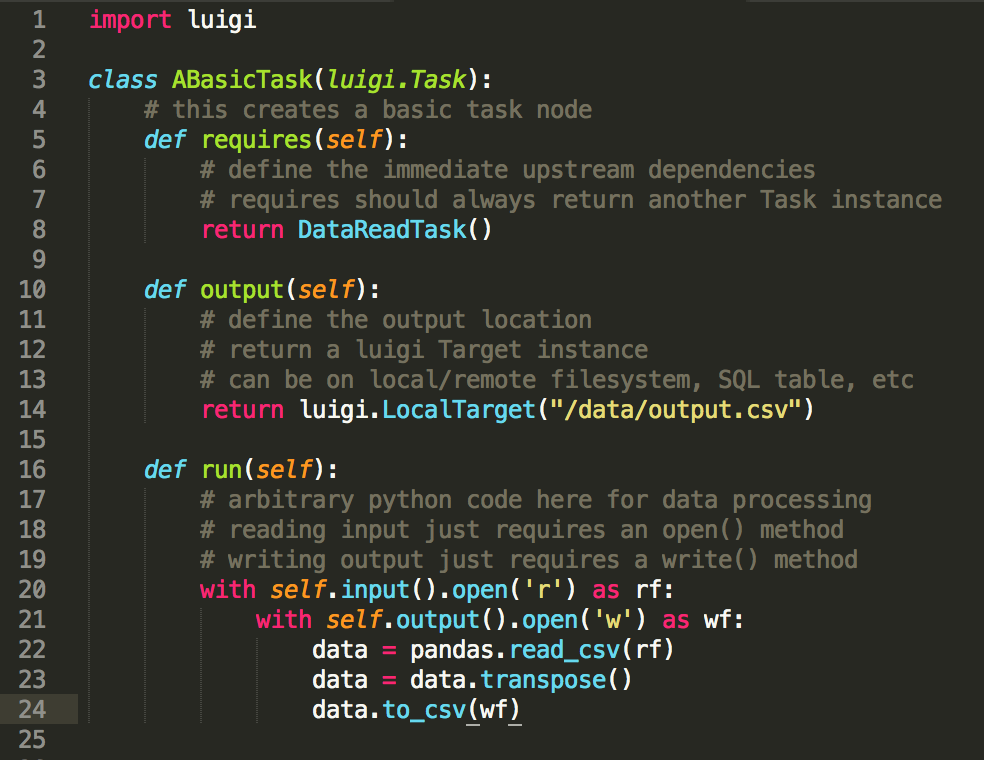
The start and end nodes of the graph are designated with special classes –
for example, the ExternalTask that points to a dependency outside of the
graph (like the raw data file) to start the pipeline only specifies the
output target for the next node(s) to ingest, while a terminal node that
triggers other tasks (the WrapperTask) only needs requires to define the
triggers.
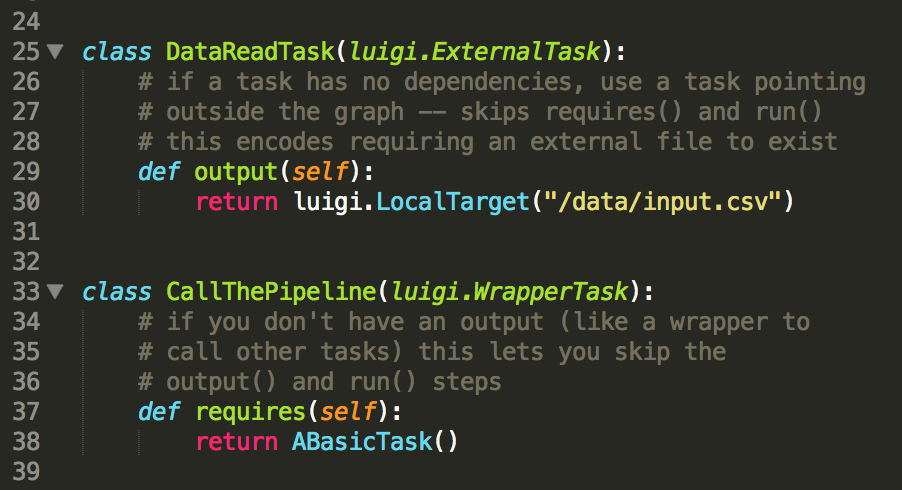
And that’s it! triggering the task manager automatically builds the graph by backtracking through dependencies from the called node, assigning workers to tasks in parallel. Repeated instances of node classes handles embarassingly-parallel tasks – the graph below was specified in less than a hundred lines of code all told.
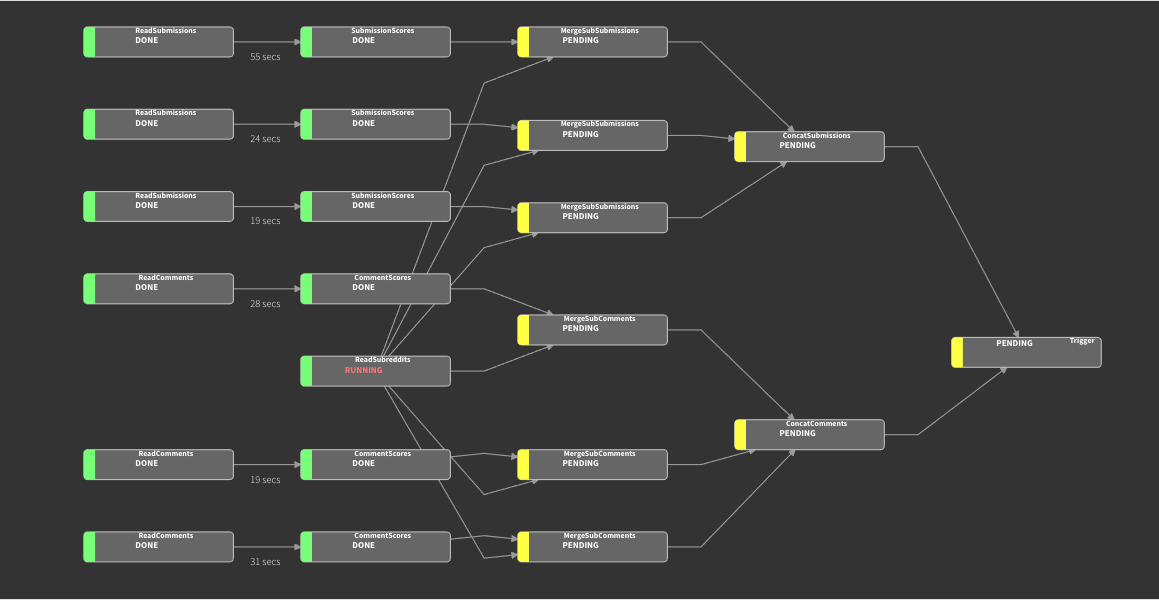
The Luigi server even generates a web interface for job monitoring for you. It doesn’t solve everything – for example, Luigi doesn’t have an included scheduler, so tasks like nightly reports will need a separate CRON call. But it allows for quite powerful task graphs, since Luigi can directly integrate to other data tools (e.g. S3, HDFS, Apache Spark) with minimal overhead or learning curve.
scalable analysis tools
While Luigi allows for multiple task workers operating in parallel, each worker is still limited by machine resources – the pipeline speed is capped by processing power, while large dataset processing is limited by machine memory. Lacking processing power makes the job slower… lacking memory means it won’t run at all! We need parallelization/distribution of computing to apply more processing power to the task, and out-of-core computing to remove the limits of doing all our work in-memory on the machine.
Fortunately, we have a number of powerful tools at our disposal: I particularly
like dask, a Python package for out-of-core
wrappers to numpy and pandas, and Apache Spark,
an enterprise-level distributed computing system for the Hadoop ecosystem.
Each has their strengths: I like dask for easy prototyping and direct Python integration:
- wrapper around
pandas-based Python workflows: drop-in replacement for ubiquitous & powerful data tools - trivial to implement at prototyping level, giving a performance boost even on single laptop
- integrated with sklearn, xgboost, tensorflow for ML tasks
- python-specific!
while spark is powerful for standing pipelines, atomic tasks, and cross-language use:
- built from the ground up for the “thousand-node cluster” use case, intended for remote connections to central cluster
- best used for primitive tasks (executed on huge datasets)
- can directly integrate with Luigi pipeline
- accessible from Python, R, Scala, Java
I frequently use both for different stages of the data pipeline. I find Spark
best suited for extremely efficient implementation of simpler transforms or
map/reduce operations across Resilient Distributed Datasets (RDDs) for
standing ETL tasks, although it also has strong support for more advanced
operations up to and including ML tasks. For more complex transforms,
I prefer dask to take advantage of the built-in advanced functionality of
Python data tools, since it directly integrates with pandas for data
manipulation (so it’s a minimal change from the original pandas workflow),
and sklearn and other ML packages for modeling.
visualization and analytics
So now that we have a healthy pipeline for our data preparation and analysis, we need to be able to present our results in a repeatable, documented way – either by visualization, or aggregated metrics. Python already has some powerful tools for this – Jupyter notebooks run Python code with embedded visuals and Markdown annotation, and visualization packages like seaborn and bokeh can easily produce compelling results. However, anything with a Jupyter notebook requires some code knowledge (at least to the level of rerunning scripts) and can be tricky to share in a formulaic way (though tools like knowledge-repo can help).
Web-based dashboards generally make for a better solution, since it is straightforward to reproduce dashboards from a centralized service laterally across teams or longitudinally with data over time. The web interface lets you democratize access to data & analytics across the group, without the code-knowledge barrier to entry.
naturally, this is one of the most crowded spaces for commercial products in the data analytics space.
There are numerous commercial solutions to dashboarding – Tableau, Chartio, Plotly, or Looker just to name a few. While excellent products, they can run to the pricier side. Fortunately, there are several open-source solutions as well! In my time at Cinch, some of the better options I encountered:
-
superset, originally created by the DS team at AirBNB and now maintained by the Apache foundation
-
metabase, a “freemium” product maintained as an open-source tool, with the developers selling product support and hosting
-
plotly, another open-sourced core tool (with paid support and hosting), along with excellent visualization packages for Python and R
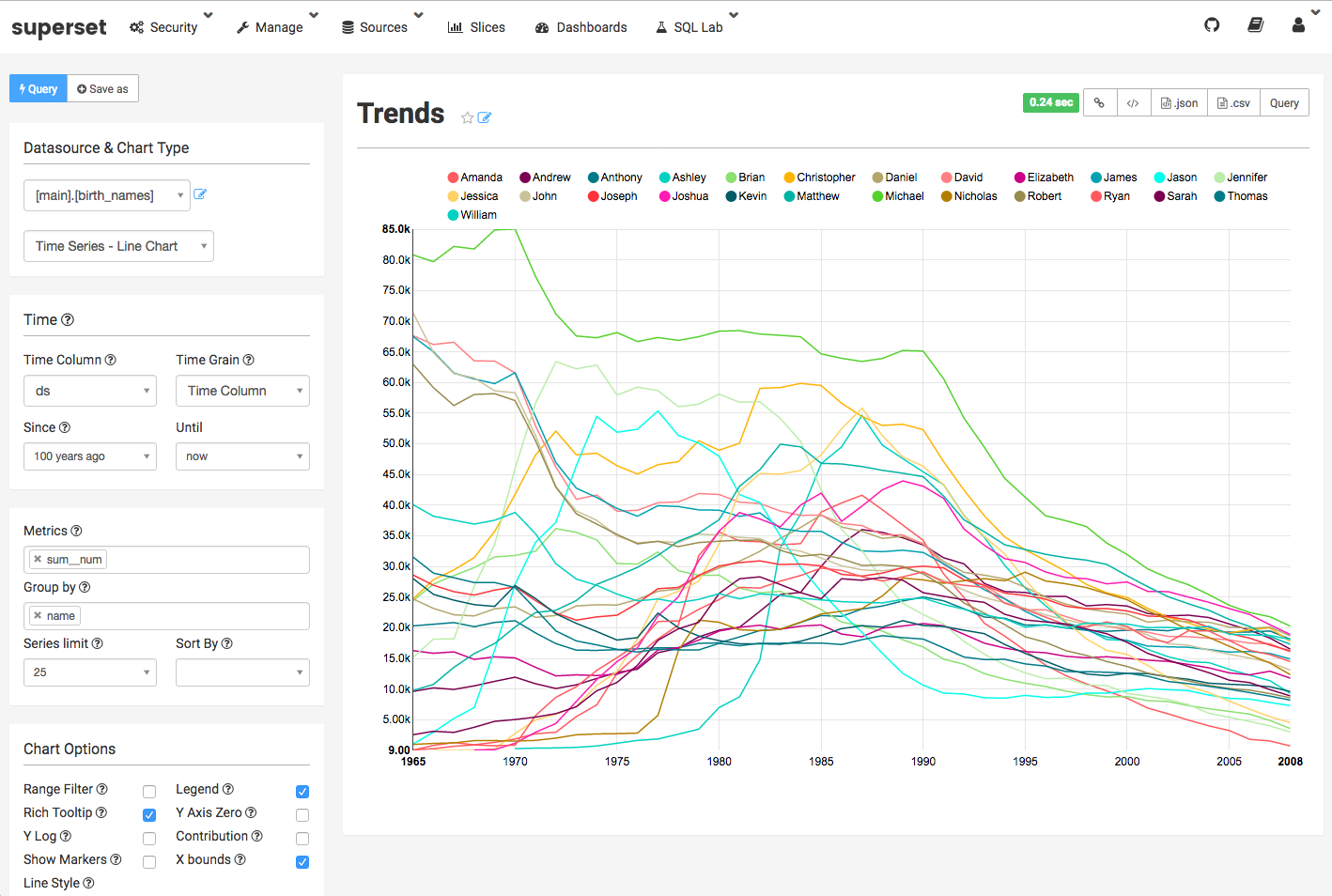
Each of these enables web-based, code-free development of dashboards, data exploration through a SQL-like interface (no SQL knowledge required, though they also support direct interaction with the SQL backend) and sharing of reproducible reports and dashboards across teams. I generally found metabase to be more visually appealing, but superset has broader open-source development.
However, each of these tools presupposes a database structure of some form to act as its backend, so they can’t operate directly off of the data lake structure. The data pipeline can be used to create and populate this staging database, though – either by regularly populating preprocessed data into a persistent OLAP database, or by creating a DB instance on the fly (suitable for smaller datasets), like in the simple local example below:

staging local data through the pipeline into a SQLite instance, and spawning a superset instance reading it. This naturally has scaling issues (DB constraints on SQLite, local hosting, on-the-fly processing for larger data) so an intermediate analytics database is a good idea for anything beyond smaller-scale prototyping.
tying it together
While you can certainly gain value from an integrated workflow even for personal projects, the real strength of these tools come in to play when working as part of a team. The centralized data store and ETL pipelines ensure everyone is looking at the same data (a seemingly simple need that often breaks down when sharing via static file dumps to Google Drive or similar), while central computing resources for Python, R, or Spark allow any member of the team to access processing power well in excess of their own local machine.
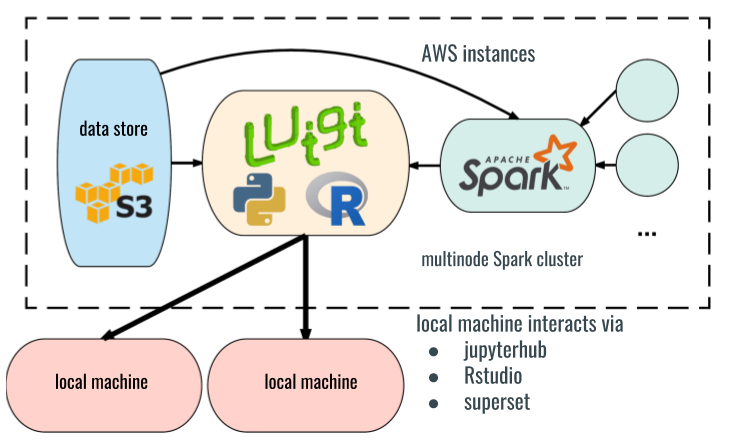
Integrating with AWS makes for efficient use of shared computing resources with efficient connections to S3 storage, and is naturally suited to sharing analysis with a broader team. Processing pipelines and analysis tools are hosted on a central hub EC2 instance, with a direct connection to S3 for data access. Analysis tools can be run without command-line/SSH access to EC2 via controlled Jupyterhub or Rstudio server access. Ancillary EC2 nodes can be spun up as needed as workers on an attached Spark cluster driven from the hub node. This ensures access to all the needed resources, with access secured behind a common VPC.
Apart from the AWS computing resources, every tool in this post can be built
with free and open-source software. The tools can largely be installed via
pip or anaconda in Python, or be found at:
| package | links |
|---|---|
| luigi | github, documentation |
| dask | github, documentation |
| seaborn | github, documentation |
| bokeh | github, documentation |
| superset | github, documentation |
| metabase | github, documentation |
| Apache spark | binaries, documentation, cluster control scripts |
| Jupyterhub | github, documentation |
| Rstudio | homepage |